One of the main research interest in our lab focuses on the evolution of centromeres - specialized chromosomal regions, which enable the assembly of the kinetochore protein complex and the attachment of spindle microtubules to ensure the faithful segregation of sister chromatids during cell division. Paradoxically to this essential function it is surprising that different strategies of centromere organization have evolved. While most eukaryotes have monocentric chromosomes where spindle attachment is restricted to a single chromosomal region resembling such classic X-shape like structures under the microscope, many lineages have evolved holocentric chromosomes where spindle microtubules attach along the entire length of the chromosome.
To understand the evolution and regulation of holocentromeres we use insects as model system. We apply microscopic, cell biological, genomic and biochemical analyses to study the organisation and function of centromeres and kinetochores in these species. See also:
Cortes-Silva et al., Current Biology, 2020
Senaratne et al., Current Biology, 2021
Sankaranarayanan et al., bioRxiv, 2024
Yu et al., bioRxiv, 2025
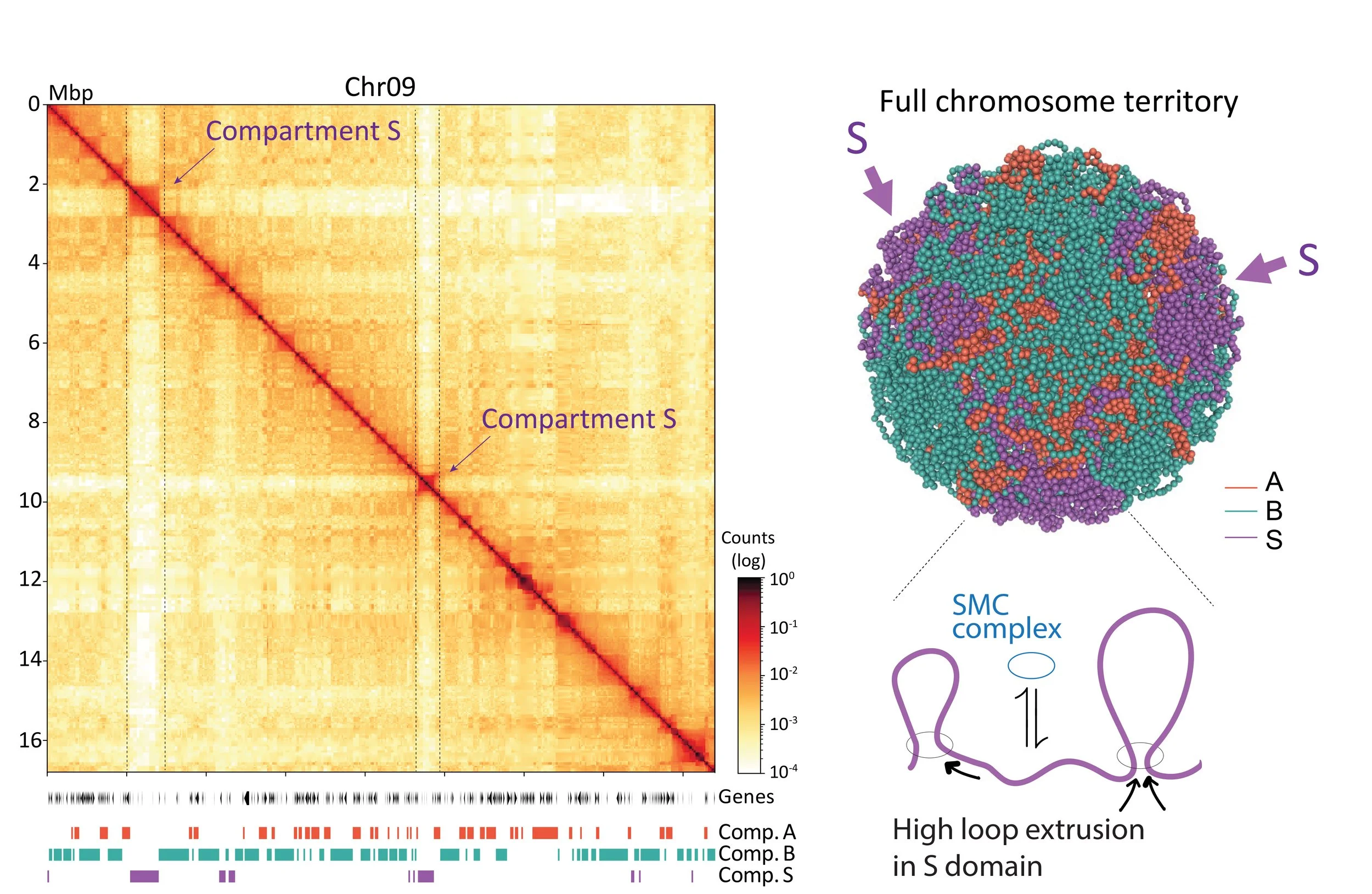
In another line of research, we aim to understand the spatial organization of holocentric chromosomes and the conservation of 3D genome architecture across Lepidoptera. To this end, we employ comparative genomic approaches in combination with microscopy and Hi-C analyses across multiple species. To learn more see Gil et al. bioRxiv, 2024